Problem Definition
Part 1: We are meant to implement one of the segmentation methods indicated in class, such as dynamic thresholding, to segment out the tumors in each image (black binary image on white background)
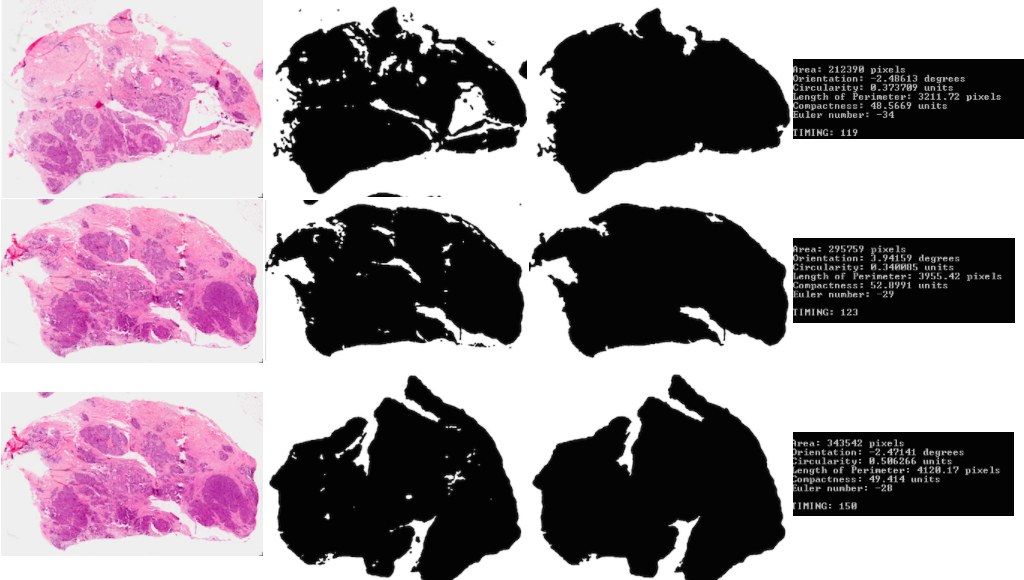
Part 2: Here we are meant to process various attributes about the binary object detected: area, orientation, circularity, length of perimeter, compactness, and the euler number.
Part 3: In this part we must implement a skeleton creating algorithm on images that had distinct streaks of more vivid red, the red tissue folds.
Part 4: Here utilize the tools at hand to try and identify an unnamed cancer tumor slide.
Part 5: Similar to the above, except now we know part of the slide is missing or damaged, the algorithm must be modified to still effectively identify the slides.
Part 6: This part asks a question of effectiveness of the algorithm for the same slides with pictures taken at different times.
Method and Implementation
Method
Part 1-2: These two parts are implemented together in the function “part1”. First, it converts the images to black and white and then runs a contours algorithm to find the biggest contour, which will be the most appropriate outline of the object. Using “findBiggestContour”, the area of the biggest contour is filled. Note that by this, we define the area of the tumor to include all of its holes and inside area of the perimeter contour.
Part 3: This implements the thinning algorithm, the functions for which are defined and explained more below. Each image is read in and then thresholded using an “average threshold” where the dividing point of the binary image threshold is the average intensity found across the image, which was found to be sufficiently effective.
Part 4: In this part, we read in the images of the database and run the algorithm of part one for each image. This gets the area, orientation, perimeter, and other such previously mentioned. Then, the image lacking ID is read in (in this case, we just read in one of the previous images unaltered) and run to find its according parameters. Then, it is compared against the parameters of each of the other images. For each category or parameter, a sum of the percentage difference in values for each is added together to a total “difference” variable. The two images with the lowest difference value matches the best.
Part 5: In this section, it was clear that the previous method would not work as is. Instead, we use template matching of the “broken” image and compare against all images in the database. We simulated the broken image by cropping and drawing extra whitespace onto some random samples. The Program reads in the template or broken file, and iterates it over all images in the database, noting each time the maximum value or match. Only the best match is used. Note that if the broken image or template is larger than the image it is being run against, we immediately know that it is not a match as the sizes differ (the potentially equal or smaller in size broken piece cannot be larger than the original).
Part 6: Considering how the previous methods are implemented, a combination of parameter matching (area, orientation, etc) and template matching would be the most effective at matching the images. The area and perimeters would likely be very similar even if the sample has aged some, meaning it would be quite effective, while any changes in shape or exact locations of the smaller tumor tissue folds would lead to significant decrease the in the effectiveness of the template matching method. Some extra thresholding consideration would be needed to consider on how to weight the value of the matching parameters and the template matching, but I expect it largely would still work.
Additional Functions
- calculateEulerNumber: This function calculates the Euler number (holes versus bodies) by running the contours algorithm twice, including and excluding additional floating holes. The appropriate difference gives the Euler number.
- convertToBinary: This function creates the binary image from a color image with some erosion and dilation to get rid of unnecessary detail and then automatically threshold it.
- findBiggestContour: This function finds the biggest contour in a set of several, by checking the longest length.
- getOrientation: This function gets the orientation of the object using the algorithm provided in class, as placed in the code.
- calculateCircularity: This calculates the circularity for the input binary image, using the Emin/Emax method described in class. This is implemented by the following function:

- thinningIteration: This is a helper function for the thinning function.
- thinning: This function implements the thinning algorithm. The basic algorithm iterates through the entire image and checks whether pixels are odd or even, and in certain predetermined conditions deletes the pixel (thins it) recursively
Experiments
The experiments and different trials can be shown below, and each with their respective timings.
Results – tumor properties
List your experimental results. Provide examples of input images and output images. If relevant, you may provide images showing any intermediate steps
Results |
||||
| Trial | Source Image | Intermediate Image | Final Image | Properties & time |
| Part 1: trial 1 |  |
 |
 |
 |
| Part 1: trial 2 |  |
 |
 |
 |
| Part 1: trial 3 |  |
 |
 |
 |
| Part 1: trial 4 |  |
 |
 |
 |
| Part 3: trial 1 |  |
 |
Time: 75299 | |
| Part 3: trial 2 |  |
 |
Time: 43608 | |
| Part 3: trial 3 |  |
 |
Time: 12012 | |
More Results |
||
| Trial | Result | |
| Part 4: trial 1 |  |
|
| Part 4: trial 2 |  |
|
| Part 4: trial 3 |  |
|
| Part 4: trial 4 |  |
|
| Part 4: trial 1 |  |
|
| Part 4: trial 2 |  |
|
| Part 4: trial 3 |  |
|
Discussion
Discuss your method and results:
- Our results are generally successful. For part 3, we used a simpler segmentation method (average), but it still worked for the most part to get the right segments. We can see the skeleton thinning works well, though in the last experiment it did not run all the way across the image as it should have.
- This program could be improved greatly in efficiency. As the timings show, the thinning program is very slow. A better algorithm would be to do sub-thinning calls for smaller, independent sections of the image with the object so that the processing recursively gets faster as it covers less area, instead of always looking at every pixel. For part 4 and matching lost ID’s with the database and with broken or aged specimens, better weighting or analysis could be done on the parameters instead of just adding the percentage difference. For example, scaling could be considered and orientation perhaps would be less useful in case the item can rotate on the slide naturally between captures.
Conclusions
My main conclusion is that thinning is a heavy algorithm that needs careful and effective segmentation to utilize properly. My other main takeaway is that it is easy to get many properties off of an image to use as faster, easier comparisons than by doing fuller/more computationally heavy processing like template matching or thinning.
Credits and Bibliography
- “Computer Vision for Interactive Computer Graphics”, handed out in class on September 16th
- http://opencv-code.com/quick-tips/implementation-of-thinning-algorithm-in-opencv/, accessed October 5, 2014.
- openCV Documentation
This homework was developed with Timothy Chong.